Summary
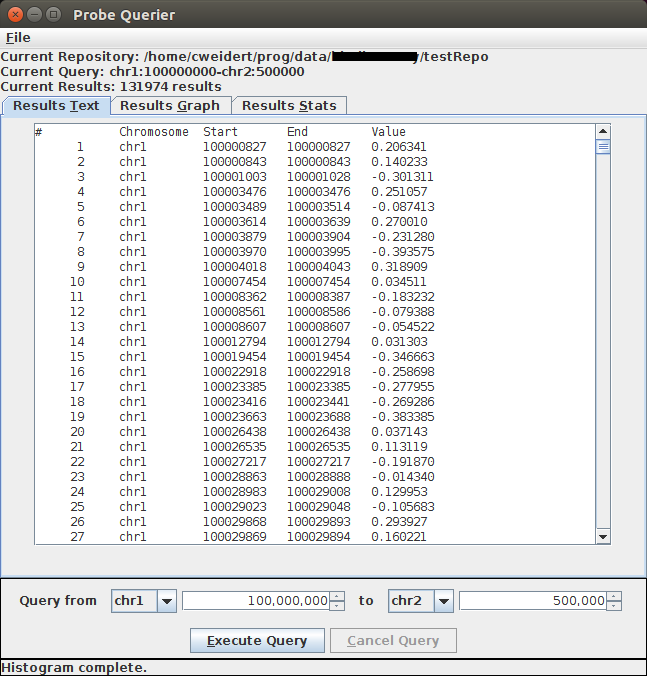
This is a project I was working on in November 2016. It's to visualize readings from individual genomes. The idea is that each reading has a start point, an end point, and a value, which indicates the strength of a match between the start and end point. I was tasked with structuring the data in such a way that a user could perform fast queries on the data and display the results. A typical query might be something like "I want to select all the readings between chromosome 1 offset 1,000,000 and chromosome 2 offset 500,000."
This program was only ever run on a toy dataset with approximately 2.9 million readings. In real life the dataset would be much larger. I was explicitly forbidden from using databases.
Results Text Display
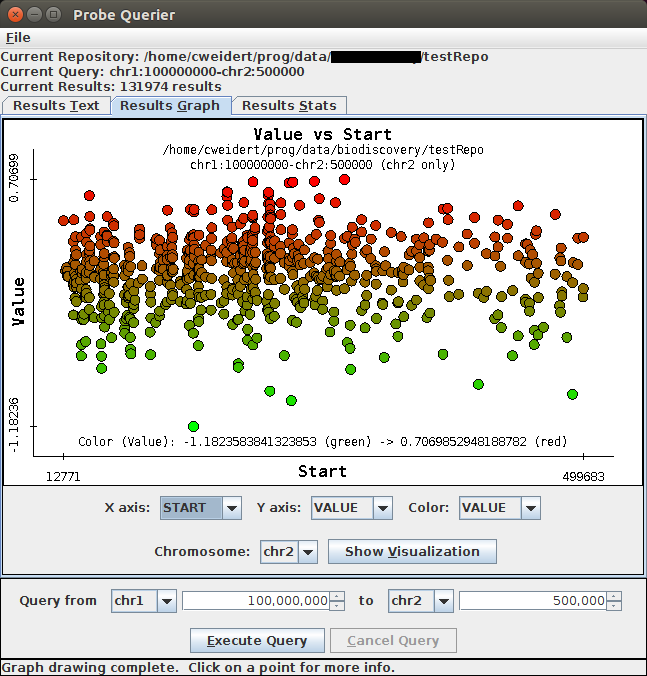
Results Graph Display
Results Graph Display
Programming Concepts Used
- I used lambda selections to extract the various measures from the readings in the results.
- I used Java's Swing package to make the GUI.
- Because I was forbidden from using any kind of database, I wrote a B-Tree to store the data in some sorted form. I somewhat arbitrarily chose each node to have a maximum of 1,024 readings. Each node would be saved to the disk for quick future loading.
- I used threads to read the data file and build the tree structure.
- I used enums, regular expressions, interval trees, interfaces, abstract classes, polymorphism, affine transforms, singletons, factory classes, and functional interfaces.